RPAN2: Rice Pan-genome Browser 2
Lab of Computational Genomics and Metagenomics, Shanghai Jiao Tong University
Institute of Crop Sciences, Chinese Academy of Agricultural Sciences
Rice is one of the most important food crops globally. In 2017, we created the Rice Pangenome Browser (RPAN) to present a linear pangenome based on approximately 3K rice genomes collected using short-read sequencing, providing a vital resource for rice research and breeding programs. Recent advancements in long-read (also known as third-generation) sequencing technologies have enabled the production of a growing number of high-quality individual genomes. These advancements enable the assembly of more complete and accurate pangenomes. Here, we introduce RPAN2 - an upgraded version of RPAN that retains all previous short-read data while integrating new pangenomes constructed from long-read data. This new version features both a linear and a graph-based pangenome, supported by enhanced visualization methods.
This database provides three pangenomes:
- 3K-RG: A linear rice pangenome constructed from short-read sequencing data of the 3K Rice Genome Project (3K-RGP), containing ~ 260 Mbp novel sequences and 15,362 novel genes.
- 111TGS-RG: A linear rice pangenome derived from third-generation sequencing (TGS) data of 111 rice accessions with 113 samples, comprising 879 Mb novel sequences and 19,319 novel genes.
- 326TGS-RG: A graph-based rice pangenome built from reference genome and 263 high-quality rice genomes assembled from long reads from three previous studies and additional publicly available resources.
3K-RG
A linear pangenome
A total of 3010 rice genomes:
1787 XI(Indica), 848 GJ(Japonica), 7 cA(circum-Aus), 6 cB(circum-Basmati), 98 Admix and 264 unknown.
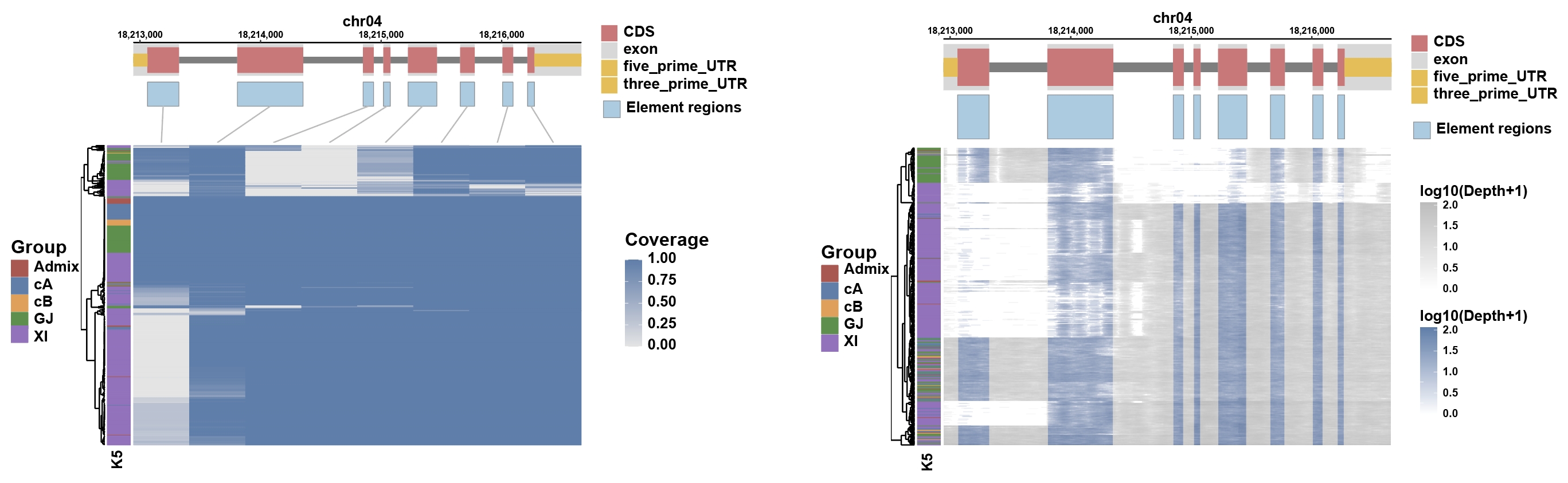
111-TGSRG
A linear pangenome
A total of 113 rice genomes:
62 XI(Indica), 25 GJ(Japonica), 9 cA(circum-Aus), 9 cB(circum-Basmati), 1 Admix and 7 unknown.
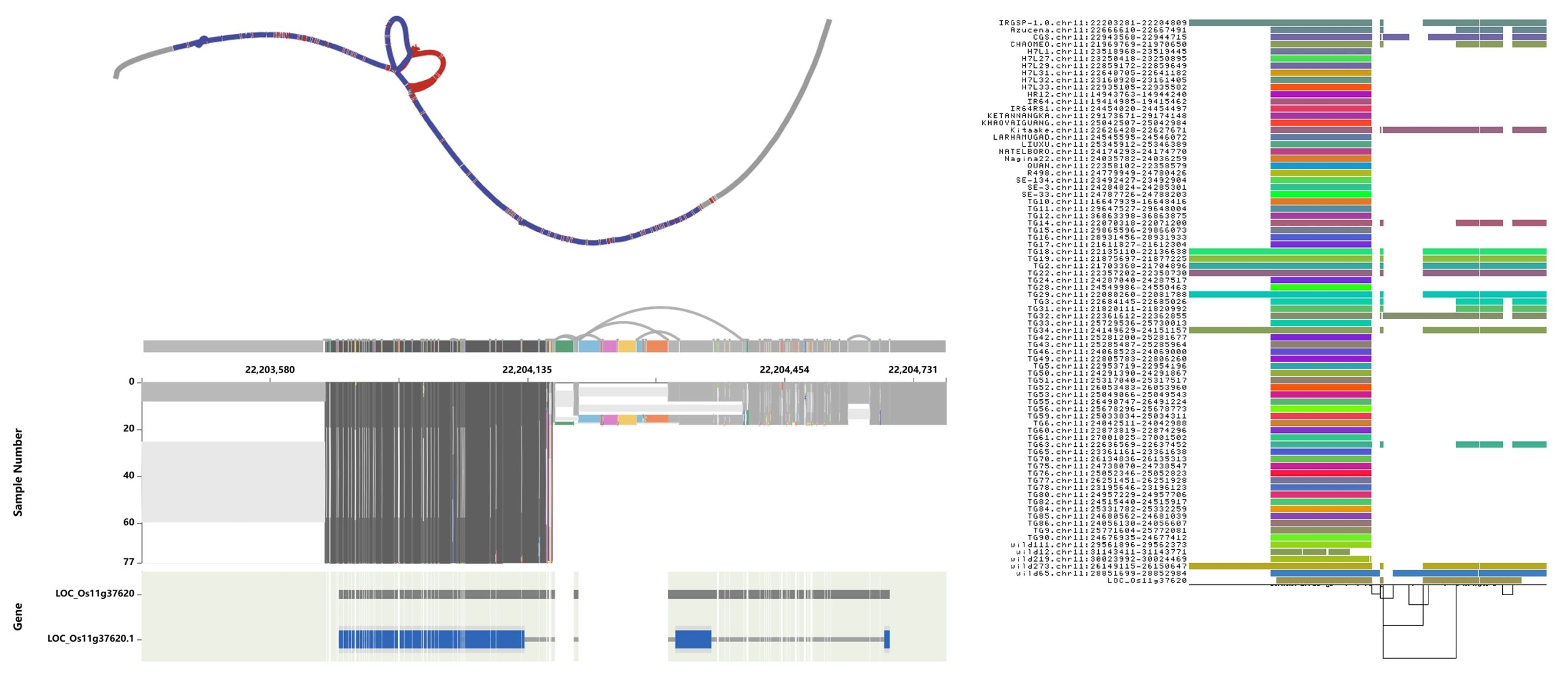
326-TGSRG
A graph-based pangenome
A total of 326 rice genomes:
222 XI(Indica), 89 GJ(Japonica), 11 cA(circum-Aus), 4 cB(circum-Basmati).